Handling missing data is a critical task in numerous fields, including data science, machine learning, and statistics, where the quality and completeness of data directly impact the performance of models and the accuracy of analyses.
In a previous tutorial, we explained how to manually handle missing data in a Pandas Dataframe. While effective, these methods often require a fair amount of coding and can be time-consuming, especially in complex datasets or scenarios.
Recently, I came across the Python feature-engine library. This powerful tool automates many of the tasks associated with handling missing data, thus streamlining the data preprocessing phase of your projects. The feature-engine library offers a suite of techniques specifically designed for feature engineering, including several innovative methods for managing missing data in Pandas DataFrames.
In this tutorial, we’ll explore how to use the feature-engine library to efficiently and effectively handle missing data in Pandas DataFrames.
Installing and Importing Required Libraries
The following script installs the libraries (including feature-engine) you need to run the codes in this article.
!pip install seaborn
!pip install pandas
!pip install numpy
!pip install feature-engineThe script below imports the libraries we just installed along with some imputation classes we’ll use for different missing data handling techniques.
import pandas as pd
import seaborn as sns
from feature_engine.imputation import(
MeanMedianImputer,
ArbitraryNumberImputer,
EndTailImputer,
CategoricalImputer,
DropMissingData)
from sklearn.pipeline import PipelineThe Sample Titanic Dataset
We’ll use the sample Titanic data from Seaborn to apply our missing data techniques. You can import the dataset using the Python seaborn library, like we do below. The output of the script confirms we have some missing values in the dataset.
dataset = sns.load_dataset('titanic')
dataset.head(10)Output:
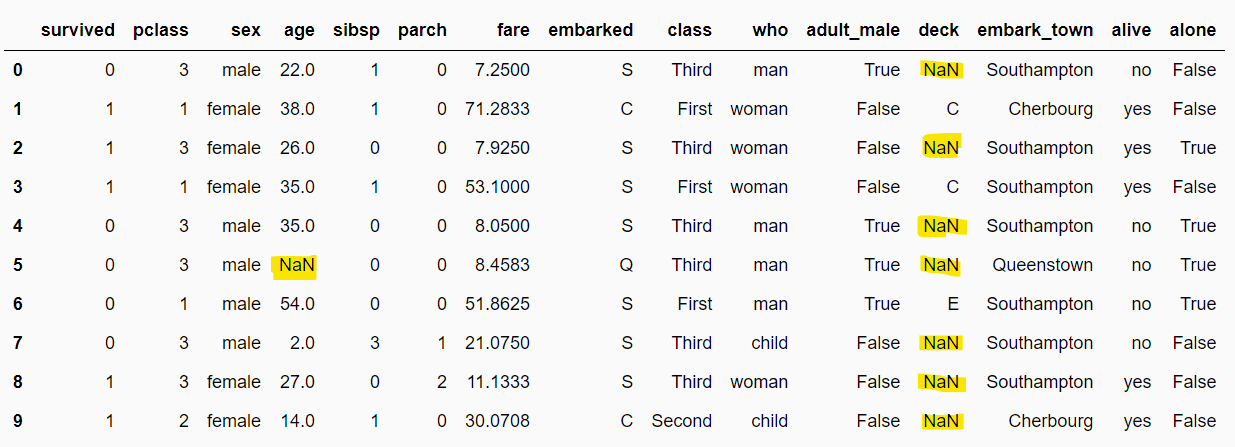
The following script displays the fraction of missing values in each column. The output shows that the age, embarked, deck, and embark_town columns are missing 19.86%, 0.2%, 77.21%, and 0.2% values, respectively.
dataset.isnull().mean()
Our dataset contains both numerical and categorical missing values, which is great since it gives us an opportunity to test the feature_engine library’s classes for handling both types of missing values.
Handling Missing Numerical Data Using Feature-Engine
The feature-engine library contains classes for the following missing data handling techniques.
- Median and Mean Imputations
- Arbitrary Value Imputation
- End of Distribution Imputation
Median and Mean Imputation
Median and mean imputations allow the missing numeric values to be replaced with the median and mean of the remaining column values, respectively.
The MeanMedianImputer class allows you to apply median and mean imputation. You must create an object of this class and pass the median or mean value to the imputation_method attribute. Next, you can call the class’s fit() method and pass your dataset to it.
The following script applies the median imputation on all numeric columns in our dataset.
median_imputer = MeanMedianImputer(imputation_method="median")
median_imputer.fit(dataset)The imputed values can be seen using the imputer_dict_ attribute. Notice that the MeanMedianImputer class automatically detects numeric columns in a dataset.
median_imputer.imputer_dict_Output:
{'survived': 0.0,
'pclass': 3.0,
'age': 28.0,
'sibsp': 0.0,
'parch': 0.0,
'fare': 14.4542}Finally, to replace the missing values with the imputed values, you can use the transform() method and pass it your original dataset.
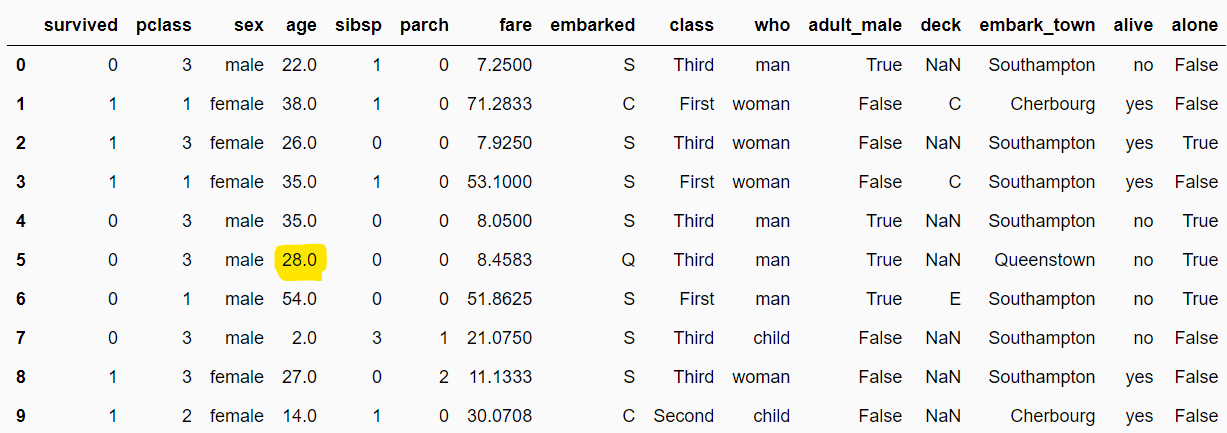
In the output, you will see that missing values in numeric columns are replaced with the imputed values. For example, the null values in the age column now all show 28.0.
filtered_dataset = median_imputer.transform(dataset)
filtered_dataset.head(10)Output:
By default, the feature-engine library applies imputation on all the columns. To apply imputations on a subset of columns, pass the column list to the variables attribute of the imputation class.
Let’s take a look. The following script handles missing values in only the age, pclass, and fare columns.
median_imputer = MeanMedianImputer(imputation_method="median",
variables = ["age", "pclass", "fare"])
median_imputer.fit(dataset)
median_imputer.imputer_dict_Output:
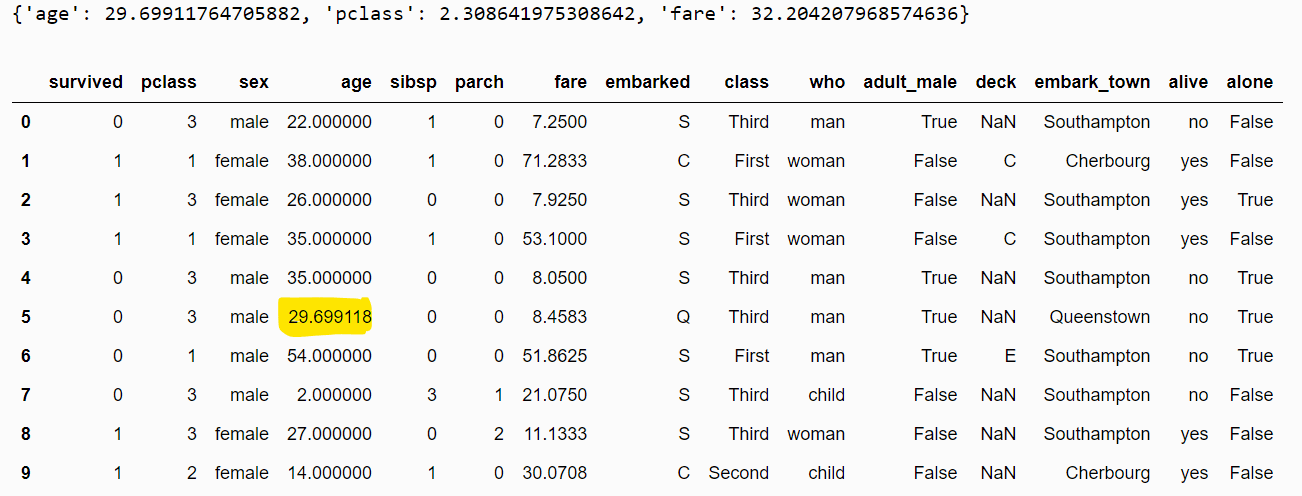
{'age': 28.0, 'pclass': 3.0, 'fare': 14.4542}Now, we know how to apply median imputation. Applying mean imputation is equally straightforward. All you must do is pass mean to the imputation_method attribute of the MeanMedianImputer class, as shown in the script below.
mean_imputer = MeanMedianImputer(imputation_method="mean",
variables = ["age", "pclass", "fare"])
mean_imputer.fit(dataset)
print(mean_imputer.imputer_dict_)
filtered_dataset = mean_imputer.transform(dataset)
filtered_dataset.head(10)Output:
Arbitrary Value Imputation
Arbitrary value imputation replaces missing values with an arbitrary number. You can use the ArbitraryNumberImputer class to apply arbitrary value imputation. You must pass the arbitrary value to the arbitrary_number attribute.
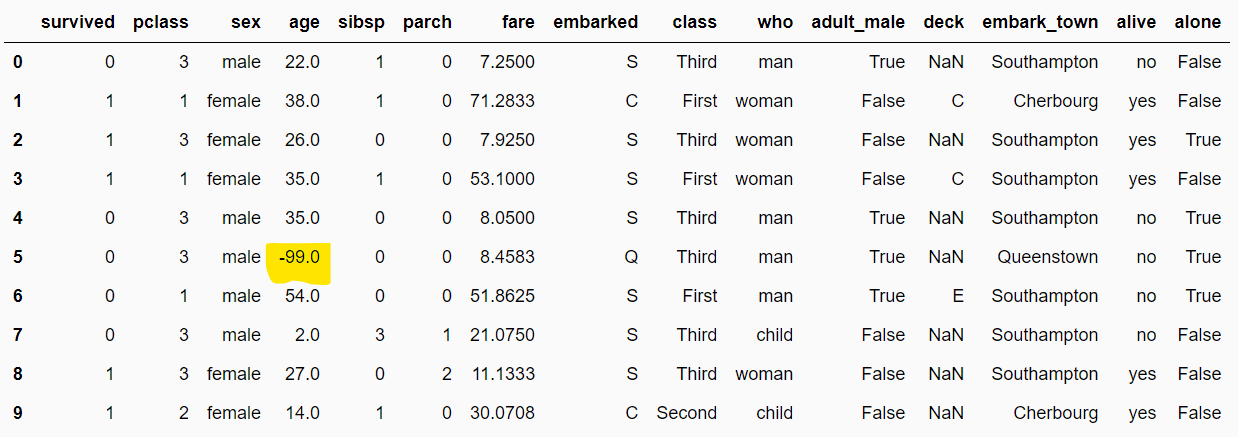
The following script replaces missing values in numerical columns by an arbitrary number -99.
arb_imputer = ArbitraryNumberImputer(arbitrary_number=-99)
arb_imputer.fit(dataset)
filtered_dataset = arb_imputer.transform(dataset)
filtered_dataset.head(10)Output:
End of Distribution Imputation
The end-of-distribution imputation replaces missing values with a value at the end of a Gaussian or inter-quartile distribution (IQR). You can use the EndTailImputer class to apply end-of-distribution imputation to missing values.
To do so, you must pass the distribution type (guassian or iqr) to the imputation_method attribute. In addition, you can pass the end of the distribution direction (right or left) to the tail attribute.
The following script selects a value from the right side of the Gaussian distributions from numeric columns and replaces it with the missing values in the corresponding columns.
eod_imputer = EndTailImputer(imputation_method="gaussian",
tail="right")
eod_imputer.fit(dataset)
print(eod_imputer.imputer_dict_)
filtered_dataset = eod_imputer.transform(dataset)
filtered_dataset.head(10)Output:
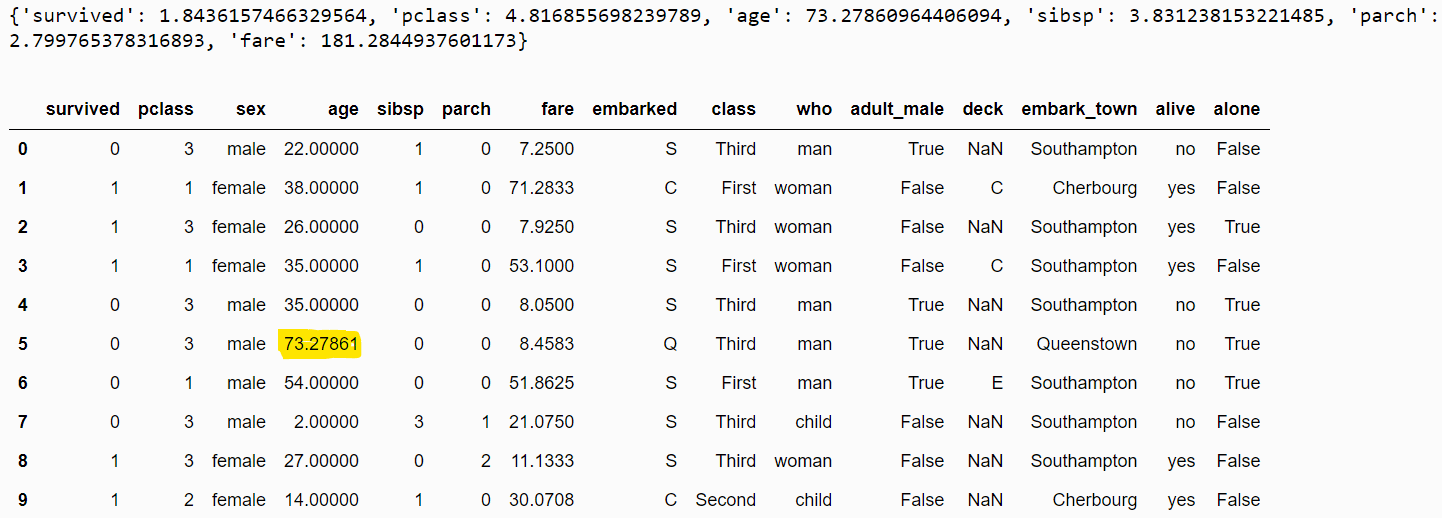
The following demonstrates an example of using inter-quartile distribution imputation using values from the left tail of the distribution.
eod_imputer = EndTailImputer(imputation_method="iqr",
tail="left")
eod_imputer.fit(dataset)
print(eod_imputer.imputer_dict_)
filtered_dataset = eod_imputer.transform(dataset)
filtered_dataset.head(10)Output:
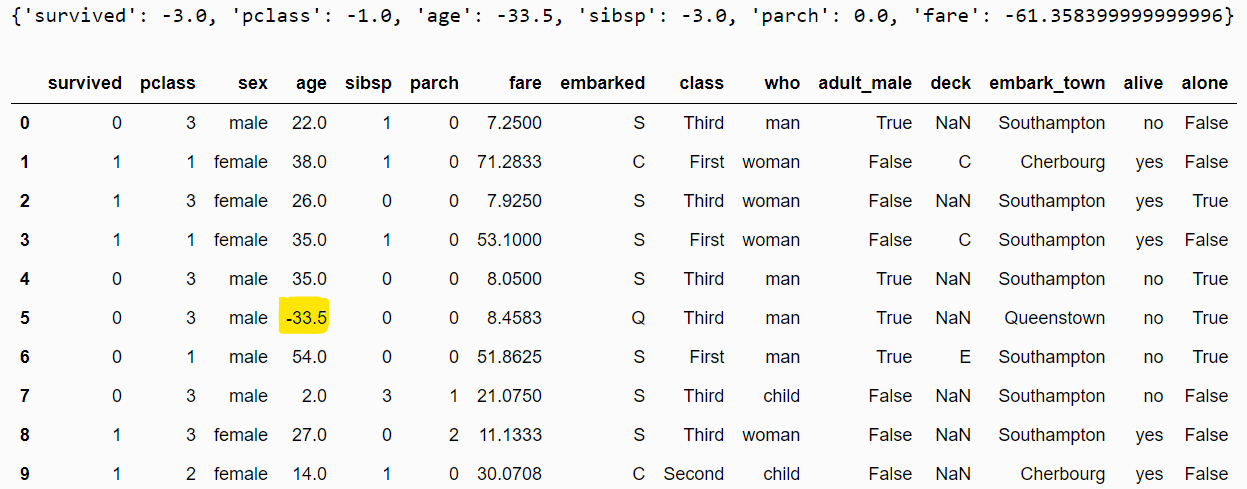
There are a variety of ways to handle missing numeric data. The examples above illustrate the importance of knowing your dataset and imputing a value that makes sense for your application.
In the next section, we’ll show you how to handle missing categorical values using the feature-engine library.
Handling Missing Categorical Data Using Feature Engine Library
The most common feature-engine techniques for handling missing categorical data are:
- Frequent Category Imputation
- Missing Value Imputation
Frequent Category Imputation
In frequent category imputation, we replace missing values in a categorical column with the most frequently occuring categories in the column.
You can use the CategoricalImputer class to apply frequent category imputation. To do so, you must pass the frequent value to the imputation_method attribute.
The following script finds the most frequently occurring categories in the categorical columns of the dataset.
fc_imputer = CategoricalImputer(imputation_method="frequent")
fc_imputer.fit(dataset)
fc_imputer.imputer_dict_Output:
{'sex': 'male',
'embarked': 'S',
'class': 'Third',
'who': 'man',
'deck': 'C',
'embark_town': 'Southampton',
'alive': 'no'}The script below replaces missing values with the most frequently occurring categories. From the output, you can see that the missing values in the deck column are replaced by the most frequently occurring category, i.e., C.
filtered_dataset = fc_imputer.transform(dataset)
filtered_dataset.head(10)Output:
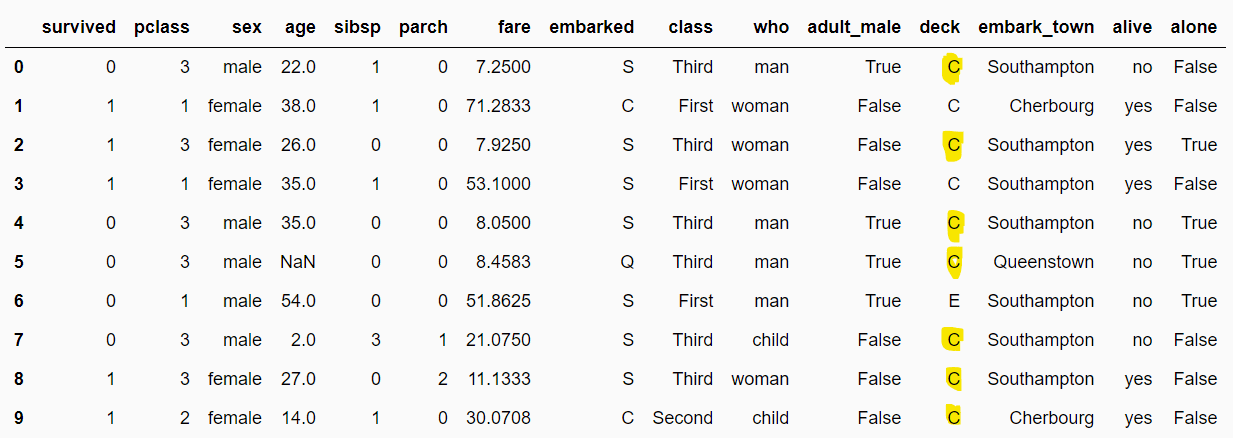
Missing Category Imputation
Missing category imputation for categorical columns is like arbitrary value imputation for numeric columns. In missing value imputation, missing values are replaced with the text Missing. You can use the CategoricalImputer class without an attribute value to implement the missing value imputation.
mc_imputer = CategoricalImputer()
mc_imputer.fit(dataset)
print(mc_imputer.imputer_dict_)
filtered_dataset = mc_imputer.transform(dataset)
filtered_dataset.head(10)Output:
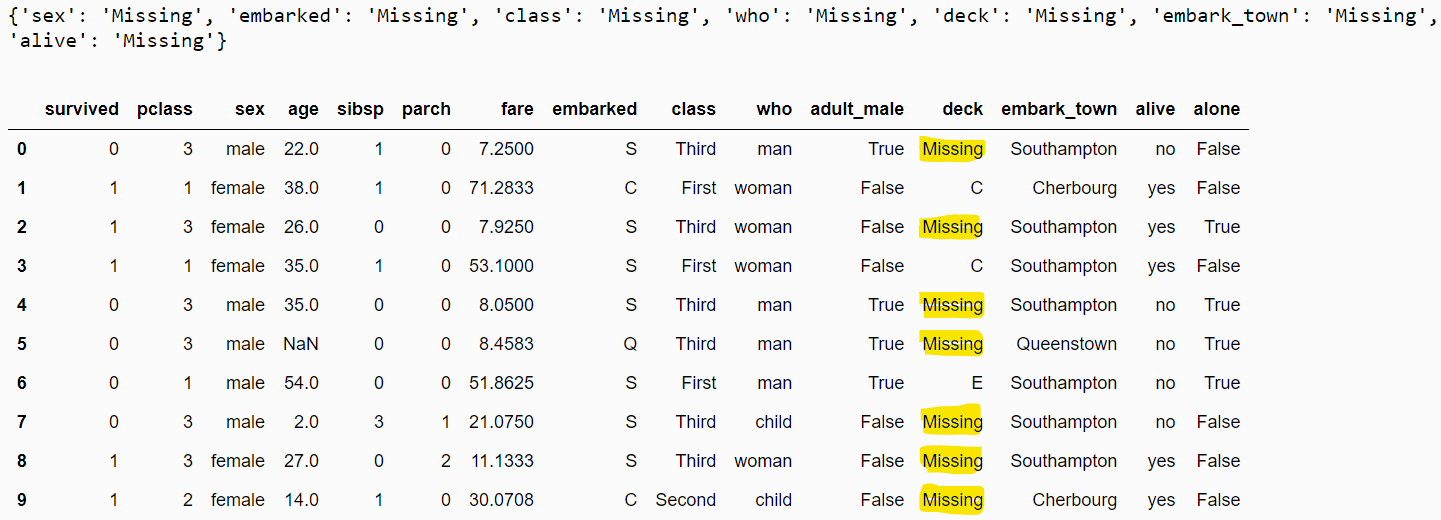
Combining Numerical and Categorical Imputations with Pipelines
One of the most exciting features of the feature-engine library is that it allows you to combine multiple imputation techniques. To do so, you must create an sklearn Pipeline and pass a list of feature-engine imputation classes for different imputation techniques.
Imputation pipelines allow you to combine numerical and categorical imputation techniques for different columns. For instance, in the following script, we apply median imputation to the numeric column
pipe = Pipeline(
[
(
"median_imputer",
MeanMedianImputer(
imputation_method="median",
variables=['age'])
),
(
"fc_imputer",
CategoricalImputer(
imputation_method="frequent",
variables=['deck'])
),
]
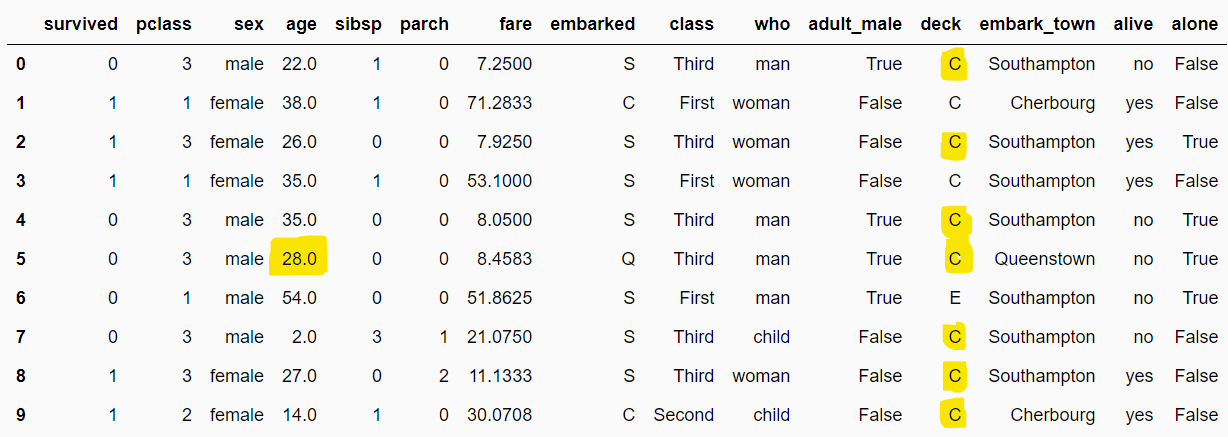
)Next, simply call the fit() and transform methods from the pipeline object to replace missing values in the age and deck columns.
pipe.fit(dataset)
filtered_dataset = pipe.transform(dataset)
filtered_dataset.head(10)Output:
Conclusion
Handling missing data is crucial for maintaining the quality of analyses and statistical models. The feature-engine library streamlines this task, offering missing data handling techniques for both numerical and categorical data.
This tutorial explained several missing data handling techniques, from basic mean and median imputations to more advanced strategies like arbitrary value and end-of-distribution imputation. By leveraging the feature-engine library, you can efficiently prepare datasets for analysis, enhancing the reliability of your results.
For more Python tips like this one, please subscribe using the form below.
